Interests
Neurodegenerative diseases
Although neurodegenerative diseases are traditionally described as protein disorders leading to amyloidosis, very recent evidence indicates that protein-RNA associations are involved in their onset. We are interested in ribonucleoprotein interactions linked to inherited intellectual disability, amyotrophic lateral sclerosis, Creutzfeuld-Jakob, Alzheimer’s, and Parkinson’s diseases. We recently focused on RNA interactions with fragile X mental retardation protein FMRP; protein sequestration caused by CGG repeats; noncoding transcripts regulated by TAR DNA-binding protein 43 TDP-43; autogenous regulation of TDP-43 and FMRP; iron-mediated expression of amyloid precursor protein APP and α-synuclein; interactions between prions and RNA aptamers. Our results are in striking agreement with experimental evidence and provide new insights in processes associated with neuronal function and disfunction.
Functional and dysfunctional ribonucleoprotein networks
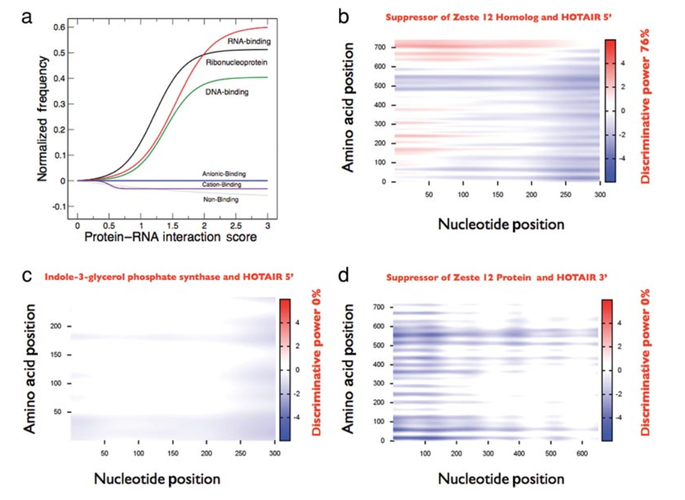
RNA-binding proteins regulate a number of cellular processes, including synthesis, folding, translocation, assembly and clearance of RNAs. Recent studies have reported that an unexpectedly large number of proteins are able to interact with RNA, but the partners of many RNA-binding proteins are still uncharacterized. Our integration of in silico and ex vivo data unraveled two major types of protein–RNA interactions, with positively correlated patterns related to cell cycle control and negatively correlated patterns related to survival, growth and differentiation. Our analysis sheds light on the role of RNA-binding proteins in regulating proliferation and differentiation processes, and we provide a data exploration tool to aid future studies
Analysis of protein networks
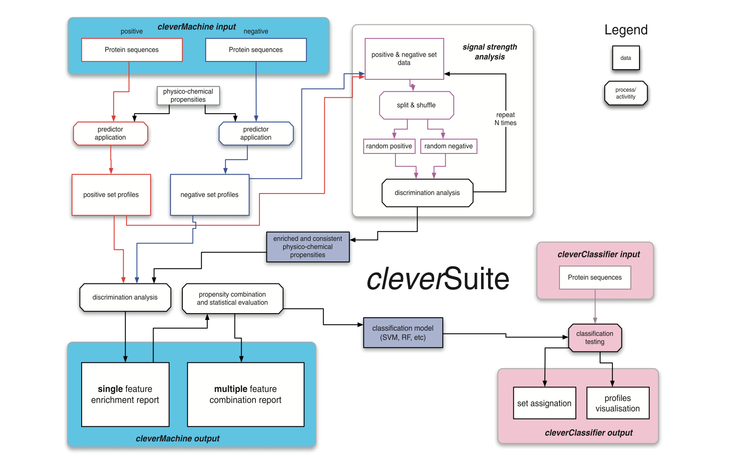
The recent shift towards high-throughput screening is posing new challenges for the interpretation of experimental results. We designed the cleverSuite approach for large-scale characterization of protein groups. The central part of the cleverSuite is the cleverMachine (CM), an algorithm that performs statistics on protein sequences by comparing their physico-chemical propensities. The second element is called cleverClassifier and builds on top of the models generated by the CM to allow classification of new datasets. We already applied the cleverSuite to predict secondary structure properties, solubility, chaperone requirements and RNA-binding abilties. Using cross-validation and independent datasets, the cleverSuite reproduces experimental findings with great accuracy and provides models that can be used for future investigations.
 tartaglialab.com
tartaglialab.com